Manual
ImageD
Interface Use:
-
Click on Begin
-
Click on File, then on Load Raw Image or Load DICOM in order to read an image to process
-
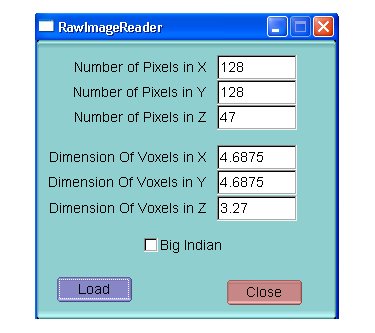
Load Raw image: Give the Image Dimensions and the pixels Size and choose Big or Little Indian (32 byte images are supported)
Or:
-
Load DICOM image
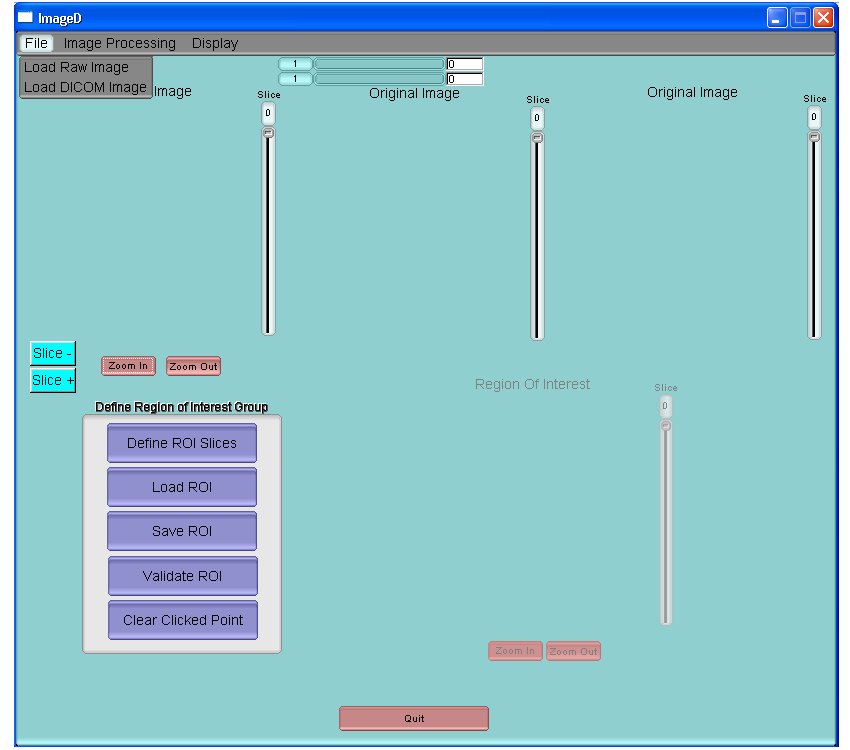
It is possible to use the variation bars above the images to adjust contrast for better visualization. Slice- and Slice+ may be used to go to the slice before or after respectively.
-
Click on Define ROI Slices to define the slices that will contain the Region of Interest around the object to process.

NB. All the subsequent mouse clicks should be carried out on the axial view only. The sagittal and coronal views are provided for display purposes only.
a. Click anywhere in the lowest slice and then click on select – Zmin
b. Click anywhere in the highest slice and then click on select – Zmax
-
Define the region of interest containing the object of interest and excluding any other significant uptake by using the mouse and clicking on 6 different points surrounding the object of interest. This contour will be copied on all the other slices that you selected on step 3. You will be able to move each point by clicking near of them in order to modify the contour in each slice if necessary.
Once the region of interest has been defined, click on Validate ROI.
You can also save or load the region of interest,. Finally, you may start over by clicking “clear clicked points” (this will erase all previously performed actions regarding the selection of slices (Zmin – Zmax) and the region of interest).
-
To get to the FLAB segmentation interface, click on Image processing FLAB Segmentation
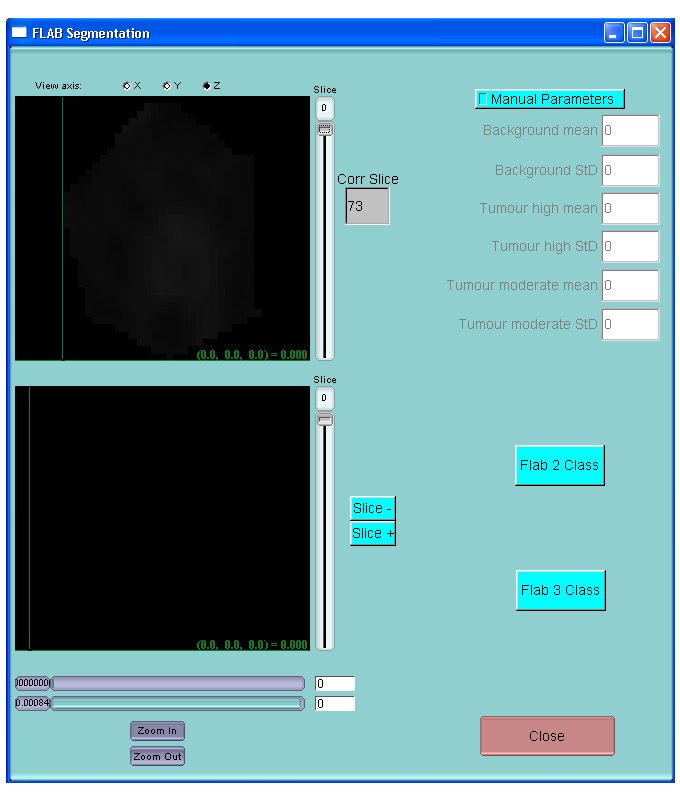
To Segment the tumor using FLAB, 2 options are available:
-
Click on Flab 2 Class for Flab using 2 classes and using automatic parameter initialization
-
Click on Flab 3 Class for Flab using 3 classes and using automatic parameter initialization
NB: FLAB 2 class should be preferred in most cases. Use Flab 3 class only for larger tumors exhibiting significant heterogeneous uptake.
For difficult cases for which initialization might fail, or if FLAB clearly results in unsatisfactory segmentation, you might click on Manual Parameter in order to specify background and tumor characteristics in order to help convergence of the segmentation algorithm. If you use 2 classes, enter values only on the first 4 rows (Background and Tumour high). If you use 3 classes, enter values in all 6 rows.
The upper figure shows the region of interest only and the lower figure shows the initial image with the integrated segmented region. It is possible to use the variation bars below to adjust contrast for better visualization.
Log file and output segmented images are generated in the same directory as the ImageD.exe file.